How to get YDNA haplogroup from AncestryDNA results
I recently learned a fun little secret about AncestryDNA tests: their tests include YDNA SNPs! It doesn't include as many SNPs as the 23andme test (885 vs 2000), but its often enough to provide men with their YDNA haplogroup. Ancestry's kit is less than half the price of a 23andme kit (currently on sale for $79 with free shipping viaAmazon.com vs. $199 for 23andme) so its a very economical method for getting this information.
AncestryDNA hides the YDNA deep within the raw data, so you have to do some digging to find the haplogroup. I wanted to make this an easier process the average genealogist could do. Here's a guide on how to get that information.
UPDATE #2 1/7/2015: I've also added a script that will automatically reformat AncestryDNA.txt to a format compatible with the 23andme to Y-SNP Converter (special thanks to redditor /u/highlandnilo). Your computer may give a virus warning when downloading it due to it being a VBS script. If this causes problems you can always follow the instructions for modifying the file yourself.
Note: YDNA data is only available for male test takers.
You may have to select the second or third choice to find your correct haplogroup. It will be the one with the most green in it. However, you should IGNORE root ancestral groups such as BT or F. If BT or F is your most likely group, proceed to the next one on the list with the most green.
To obtain your haplogroup, you'll need to run the raw data file through Promethease.com. This service reads your DNA test results and outputs a health report, all for the very low cost of $5 (there's also a slower free version available here). Simply upload the file you received from AncestryDNA to the Promethease website and it will provide you a health report. Just remember to save the report to your computer after you run it, as it will expire after 30 days!
Promethease reports categorize your results based on different medical or information topics. To find the haplogroup info, click on the "Topics" drop-down menu on the right side and select "Haplogroups." Next, select the drop-down that says "Sort by magnitude" and change this to "Sort by frequency."
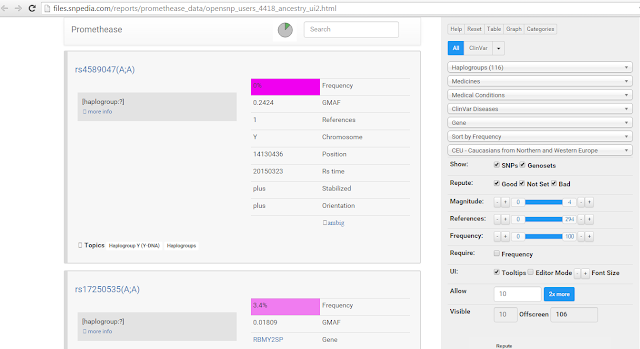
Want to see what a Promethease report looks like before you buy it? Try the above search on this example report.
You're now looking at a list of YDNA SNPs known to indicate a person's haplogroup. The top results will be the rarest, and most likely to indicate the terminal haplogroup. Unfortunately the information provided by Promethease doesn't go into detail on what each YDNA marker means. So we'll have to look at a second website to interpret the results. In a separate browser window, open up the ISOGG YDNA SNP Index. Now highlight the RefSNP ID (aka "rs" number) of the top (rarest) result in Promethease. On the ISOGG page, do a CTRL-F search and paste in the rs number. Hopefully it will be in this database and jump to the correct SNP.
On the ISOGG page, each SNP entry will have something in the rightmost column like "A->T" or "G->A". This is the change that indicates whether or not someone is positive for that particular haplogroup. The letter on the left is the ancestral version. The letter on the right is the mutation that signals your test belongs in that haplogroup. You'll want to compare this to your Promethease results to see if you are positive or negative for each haplogroup indicator.
So for example, if my top Promethease result for haplogroups is "rs17250535(A;A)" then I will search for "rs17250535" on the ISOGG page. I then see the mutation for this SNP is T->A. Since my result in Promethease was (A;A), I am positive for this mutation and belong to this haplogroup, which the ISOGG page reveals is R1a. Had I been negative (T;T), then I would have proceeded to the next SNP in Promethease. Its a good idea to try a few of the SNPs, just to be sure. Keep in mind that if you fall in a subgroup like R1a, you should be positive for all parent groups as well, such as R1 and R.
Promethease does not provide a complete listing of relevant YDNA SNPs. But typically it will provide enough for you to discover the rough haplogroup. Once you've found your group, it might be a good idea to search for more SNPs relevant to that particular group in your raw data from AncestryDNA. Admittedly its not the easiest process, but its cheaper than any of the other options for obtaining YNDA results.
AncestryDNA hides the YDNA deep within the raw data, so you have to do some digging to find the haplogroup. I wanted to make this an easier process the average genealogist could do. Here's a guide on how to get that information.
Getting a YDNA Haplogroup from AncestryDNA Results
UPDATE #1 1/6/2015: I've updated this post with a new method that uses Felix Immanuel's tools over at y-str.org and is much more automated. However, it does not use the latest version of the Y-Tree to determine SNPs (it uses 2014 version) and might have some outdated results. Its still best to check against the ISOGG Y-DNA Tree in case of changes.UPDATE #2 1/7/2015: I've also added a script that will automatically reformat AncestryDNA.txt to a format compatible with the 23andme to Y-SNP Converter (special thanks to redditor /u/highlandnilo). Your computer may give a virus warning when downloading it due to it being a VBS script. If this causes problems you can always follow the instructions for modifying the file yourself.
Note: YDNA data is only available for male test takers.
Method One: Automated SNP extraction
Step One: Download your raw data.
On the DNA Home Page, click on "Settings" on the right side of the page. On the settings page you'll see a box on the right titled "Actions." The first option is "Download your raw DNA data." Click the button just below this that says "Get Started." You'll be prompted to enter your password. After this, an email with a special link will be sent to your account's email address. It may take a few hours for this email to arrive. When it does, click the link and log in to your account. You'll then be able to download your test results. The result is a very large text file called "AncestryDNA." Save this to your computer.Step Two: Reformat the file
The program we are going to use is designed for 23andme results, so we'll need to reformat the file a bit before it will accept the AncestryDNA file as a valid input. Here is a simple VBS script that will do the reformatting for you:
Download DNAConverter.vbs.
Just download that and place it in the same directory as your AncestryDNA.txt. Double click the script and it will create a new file called AncestryDNA_Edited.txt. Now proceed to Step Three.
If you are unable to use the above VBS script, you can make the changes manually in a spreadsheet program. First, open up the AncestryDNA.txt file in Excel or your spreadsheet program of choice. You'll want to import it as a tab-deliminated file. Next we'll need to make the following four changes:
Download DNAConverter.vbs.
Just download that and place it in the same directory as your AncestryDNA.txt. Double click the script and it will create a new file called AncestryDNA_Edited.txt. Now proceed to Step Three.
If you are unable to use the above VBS script, you can make the changes manually in a spreadsheet program. First, open up the AncestryDNA.txt file in Excel or your spreadsheet program of choice. You'll want to import it as a tab-deliminated file. Next we'll need to make the following four changes:
1) Remove the header - this is the first 16 lines, all starting with a #. Delete these rows so the table headers are the first row.
2) Delete the entire Column E ("allele2").
3) Change "allele1" to "genotype"
4) Replace all mentions of Chromosome "24" with "Y". In Excel you can quickly do this by doing CTRL-F, going to the "Replace" tab, then putting 24 in as the text to find and Y as the text to replace it with. Important: Also click the options button and select "Match entire cell contents." The click "Replace All"
When finished, save this edited file as a txt file. Do not worry about any compatibility warnings Excel might give you when saving.
Step Three: Download Felix Immanuel's 23andMe to Y-SNP Converter
Felix Immanuel's Y-STR.org site has a ton of great tools for in-depth analysis of DNA results. This particular tool parses the data of a 23andme raw data file and outputs YDNA SNPs. In the last step, we converted our AncestryDNA file to the proper format for this program to read.
Run the program and select the edited AncestryDNA file you created in the prior step. It will output a list of YSNPs:
Now go to File > Save Y-SNPs. This will create a text file called YSNPS.txt
Step Four: Obtain your Haplogroup
Now go to Chris Morley's Y-SNP Subclade predictor website. Copy the contents of the YSNPS.txt file into the box on that page and click Predict. Your possible Haplogroups will be displayed on the left. It should look something like this:
You may have to select the second or third choice to find your correct haplogroup. It will be the one with the most green in it. However, you should IGNORE root ancestral groups such as BT or F. If BT or F is your most likely group, proceed to the next one on the list with the most green.
Method Two: Promethease
Step One: Download your raw data.
On the DNA Home Page, click on "Settings" on the right side of the page. On the settings page you'll see a box on the right titled "Actions." The first option is "Download your raw DNA data." Click the button just below this that says "Get Started." You'll be prompted to enter your password. After this, an email with a special link will be sent to your account's email address. It may take a few hours for this email to arrive. When it does, click the link and log in to your account. You'll then be able to download your test results. The result is a very large text file called "AncestryDNA." Save this to your computer.Step Two: Obtaining the YDNA data.
To obtain your haplogroup, you'll need to run the raw data file through Promethease.com. This service reads your DNA test results and outputs a health report, all for the very low cost of $5 (there's also a slower free version available here). Simply upload the file you received from AncestryDNA to the Promethease website and it will provide you a health report. Just remember to save the report to your computer after you run it, as it will expire after 30 days!
Promethease reports categorize your results based on different medical or information topics. To find the haplogroup info, click on the "Topics" drop-down menu on the right side and select "Haplogroups." Next, select the drop-down that says "Sort by magnitude" and change this to "Sort by frequency."

Want to see what a Promethease report looks like before you buy it? Try the above search on this example report.
Step Three: Interpreting the results.
You're now looking at a list of YDNA SNPs known to indicate a person's haplogroup. The top results will be the rarest, and most likely to indicate the terminal haplogroup. Unfortunately the information provided by Promethease doesn't go into detail on what each YDNA marker means. So we'll have to look at a second website to interpret the results. In a separate browser window, open up the ISOGG YDNA SNP Index. Now highlight the RefSNP ID (aka "rs" number) of the top (rarest) result in Promethease. On the ISOGG page, do a CTRL-F search and paste in the rs number. Hopefully it will be in this database and jump to the correct SNP.
On the ISOGG page, each SNP entry will have something in the rightmost column like "A->T" or "G->A". This is the change that indicates whether or not someone is positive for that particular haplogroup. The letter on the left is the ancestral version. The letter on the right is the mutation that signals your test belongs in that haplogroup. You'll want to compare this to your Promethease results to see if you are positive or negative for each haplogroup indicator.
So for example, if my top Promethease result for haplogroups is "rs17250535(A;A)" then I will search for "rs17250535" on the ISOGG page. I then see the mutation for this SNP is T->A. Since my result in Promethease was (A;A), I am positive for this mutation and belong to this haplogroup, which the ISOGG page reveals is R1a. Had I been negative (T;T), then I would have proceeded to the next SNP in Promethease. Its a good idea to try a few of the SNPs, just to be sure. Keep in mind that if you fall in a subgroup like R1a, you should be positive for all parent groups as well, such as R1 and R.
Promethease does not provide a complete listing of relevant YDNA SNPs. But typically it will provide enough for you to discover the rough haplogroup. Once you've found your group, it might be a good idea to search for more SNPs relevant to that particular group in your raw data from AncestryDNA. Admittedly its not the easiest process, but its cheaper than any of the other options for obtaining YNDA results.



1 comment:
Thanks for the excellent post about DNA testing to be in the best progress and for also shared valuable information for our health.
23andme login
Rhonda Patrick
warrior gene
comt gene
23andme health
Post a Comment